Plotting stats for cFOS whole-brain data sample#
The following notebokk details how to generate plots from cell statistics data.
Load data#
from pathlib import Path
import sys
sys.path.append("../..")
from napari_cellseg3d.code_models.instance_segmentation import volume_stats
from tifffile import imread
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from matplotlib.colors import Normalize
import numpy as np
import pandas as pd
path = Path.home() / "Desktop/Code/CELLSEG_BENCHMARK//RESULTS/WNET OTHERS/Seb cFOS"
labels = imread(path / "instance_labels_test.tif")
0 invalid sphericities were set to NaN. This occurs for objects with a volume of 1 pixel.
Compute statistics#
stats = volume_stats(labels)
stats_df = pd.DataFrame(stats.get_dict())
stats_df
| Volume | Centroid x | Centroid y | Centroid z | Sphericity (axes) | Image size | Total image volume | Total object volume (pixels) | Filling ratio | Number objects | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 42.800000 | 13.000000 | 0.600000 | NaN | (60, 100, 200) | 1200000 | 22515 | 0.018763 | 983 |
| 1 | 26 | 8.153846 | 15.384615 | 0.500000 | 0.830680 | |||||
| 2 | 9 | 5.000000 | 39.666667 | 0.222222 | 0.955455 | |||||
| 3 | 31 | 41.290323 | 66.870968 | 0.677419 | 0.914160 | |||||
| 4 | 4 | 7.500000 | 68.000000 | 0.500000 | NaN | |||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 978 | 12 | 33.916667 | 78.166667 | 197.916667 | 0.976335 | |||||
| 979 | 7 | 22.000000 | 7.428571 | 198.857143 | 0.861337 | |||||
| 980 | 21 | 51.047619 | 24.523810 | 198.809524 | 0.699076 | |||||
| 981 | 4 | 18.500000 | 53.500000 | 199.000000 | NaN | |||||
| 982 | 28 | 19.071429 | 91.678571 | 198.428571 | 0.880467 |
983 rows × 10 columns
stats_df["Number objects"].values[0]
983
Plottting data#
Static version#
def plot_data(data, x_inv=False, y_inv=False, z_inv=False):
data_nona = data.dropna()
x = data["Centroid x"].values
y = data["Centroid y"].values
z = data["Centroid z"].values
x = np.floor(x)
y = np.floor(y)
z = np.floor(z)
maxsph = max(data_nona["Sphericity (axes)"])
minsph = min(data_nona["Sphericity (axes)"])
cmap = cm.jet.reversed()
norm = Normalize(minsph, maxsph)
col = [cmap(norm(sph)) for sph in data["Sphericity (axes)"]]
fig = plt.figure(figsize=(15, 15))
ax = fig.add_subplot(projection="3d")
scatter = ax.scatter(x, y, z, marker="o", c=col, s=data["Volume"])
fig.colorbar(
cm.ScalarMappable(norm=norm, cmap=cmap), shrink=0.5, label="Sphericity"
)
handles, labels = scatter.legend_elements(prop="sizes", alpha=0.6)
legend2 = ax.legend(
handles, labels, loc="center left", title="Volume", labelspacing=1.7
)
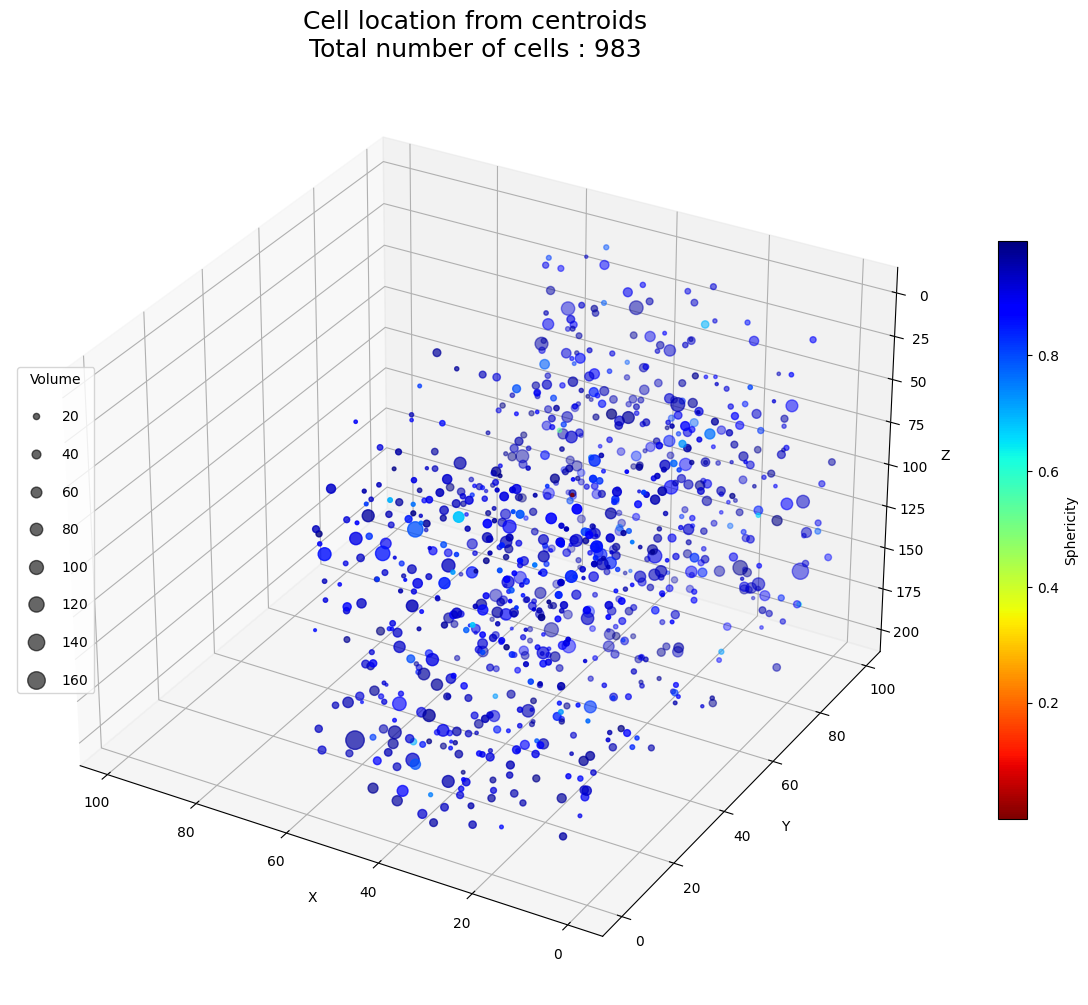
ax.set_title(
f"Cell location from centroids\nTotal number of cells : {int(data['Number objects'].values[0])}",
fontdict={"fontsize": 18},
)
ax.set_xlabel("X")
ax.set_ylabel("Y")
ax.set_zlabel("Z")
if x_inv:
ax.invert_xaxis()
if y_inv:
ax.invert_yaxis()
if z_inv:
ax.invert_zaxis()
plot_data(stats_df, x_inv=True, y_inv=True, z_inv=True)
Interactive version#
Plotly library
pip install plotlyInstallation of jupyter extensions, see Getting started > Jupyter Lab support
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode
def plotly_cells_stats(data):
init_notebook_mode() # initiate notebook for offline plot
x = data["Centroid x"]
y = data["Centroid y"]
z = data["Centroid z"]
fig = go.Figure(
data=go.Scatter3d(
x=np.floor(x),
y=np.floor(y),
z=np.floor(z),
mode="markers",
marker=dict(
sizemode="diameter",
sizeref=30,
sizemin=20,
size=data["Volume"],
color=data["Sphericity (axes)"],
colorscale="Turbo_r",
colorbar_title="Sphericity",
line_color="rgb(140, 140, 170)",
),
)
)
fig.update_layout(
height=800,
width=800,
title=f'Total number of cells : {int(data["Number objects"][0])}',
)
fig.show()
plotly_cells_stats(stats_df)